Redshift
PRODFeature List
✓ Metadata
✓ Query Usage
✓ Data Profiler
✓ Data Quality
✓ dbt
✓ Lineage
✓ Column-level Lineage
✓ Stored Procedures
✓ Sample Data
✓ Reverse Metadata (Collate Only)
✓ Auto-Classification
✕ Owners
✕ Tags
- Requirements
- Metadata Ingestion
- Query Usage
- Data Profiler
- Data Quality
- Lineage
- dbt Integration
- Enable Security
- Troubleshooting
- Reverse Metadata
Requirements
Metadata
Redshift user must grantSELECT privilege on table SVV_TABLE_INFO to fetch the metadata of tables and views. For more information visit here.
The Redshift connector supports both Amazon Redshift Provisioned (cluster) and Amazon Redshift Serverless starting from release 1.11.5. The connector automatically detects the deployment type during ingestion and uses the appropriate system views for query and lineage extraction. No additional configuration changes are required.
Profiler & Data Quality
Executing the profiler workflow or data quality tests, will require the user to haveSELECT permission on the tables/schemas where the profiler/tests will be executed. More information on the profiler workflow setup can be found here and data quality tests here.
Usage & Lineage
For the usage and lineage workflow, the user will needSELECT privilege on STL_QUERY table. You can find more information on the usage workflow here and the lineage workflow here.
Metadata Ingestion
Visit the Services Page
Click `Settings` in the side navigation bar and then `Services`. The first step is to ingest the metadata from your sources. To do that, you first need to create a Service connection first. This Service will be the bridge between OpenMetadata and your source system. Once a Service is created, it can be used to configure your ingestion workflows.

Name and Describe your Service
Provide a name and description for your Service.
Service Name
OpenMetadata uniquely identifies Services by their **Service Name**. Provide a name that distinguishes your deployment from other Services, including the other Redshift Services that you might be ingesting metadata from. Note that when the name is set, it cannot be changed.
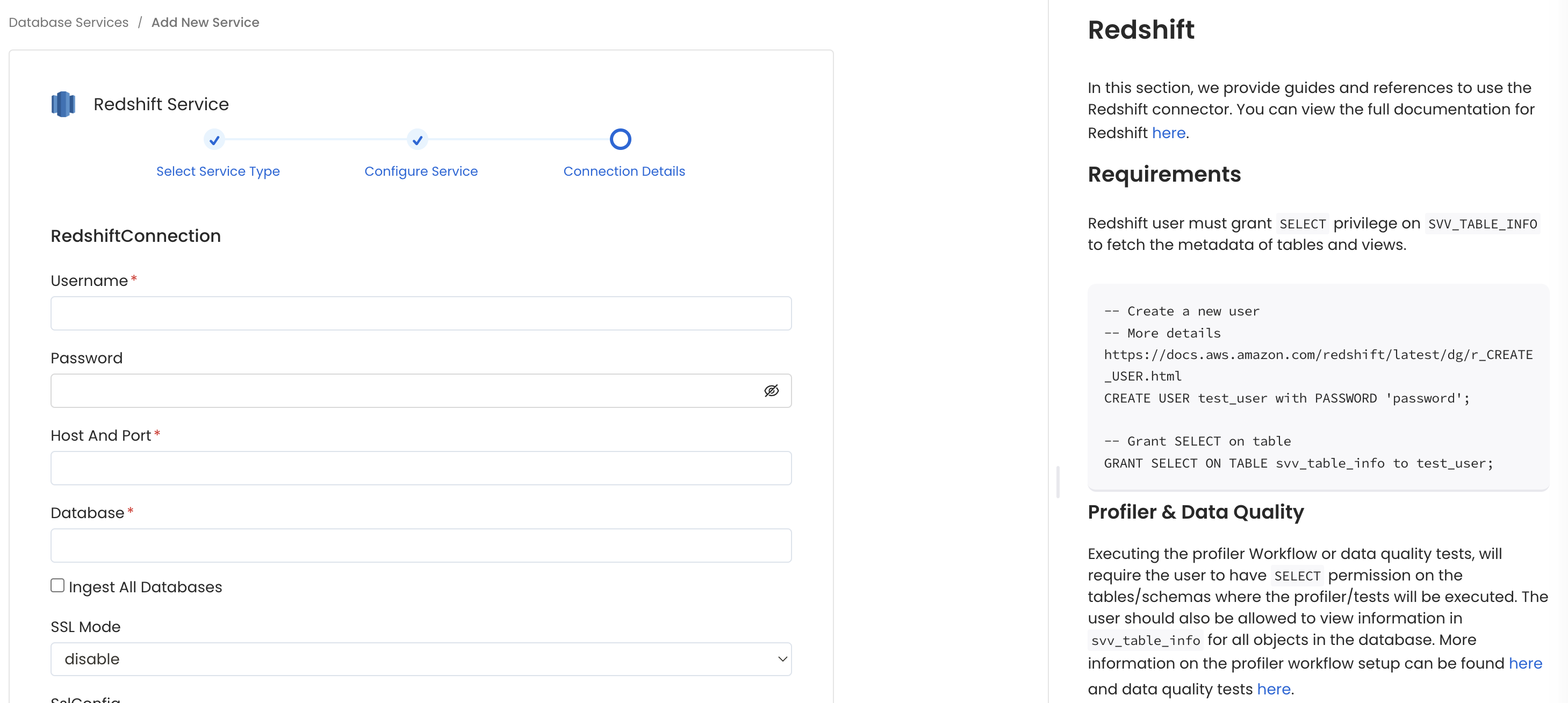
Connection Details
Connection Details
- Username: Specify the User to connect to Redshift. It should have enough privileges to read all the metadata.
- Password: Password to connect to Redshift.
- Database: The database of the data source is an optional parameter, if you would like to restrict the metadata reading to a single database. If left blank, OpenMetadata ingestion attempts to scan all the databases.
- disable: SSL is disabled and the connection is not encrypted.
- allow: SSL is used if the server requires it.
- prefer: SSL is used if the server supports it. Amazon Redshift supports SSL, so SSL is used when you set sslmode to prefer.
- require: SSL is required.
- verify-ca: SSL must be used and the server certificate must be verified.
- verify-full: SSL must be used. The server certificate must be verified and the server hostname must match the hostname attribute on the certificate. For more information, you can visit Redshift SSL documentation
Advanced Configuration
Database Services have an Advanced Configuration section, where you can pass extra arguments to the connector
and, if needed, change the connection Scheme.This would only be required to handle advanced connectivity scenarios or customizations.
- Connection Options (Optional): Enter the details for any additional connection options that can be sent to database during the connection. These details must be added as Key-Value pairs.
-
Connection Arguments (Optional): Enter the details for any additional connection arguments such as security or protocol configs that can be sent during the connection. These details must be added as Key-Value pairs.
Test the Connection
Once the credentials have been added, click on Test Connection and Save the changes.

Configure Metadata Ingestion
In this step we will configure the metadata ingestion pipeline,
Please follow the instructions below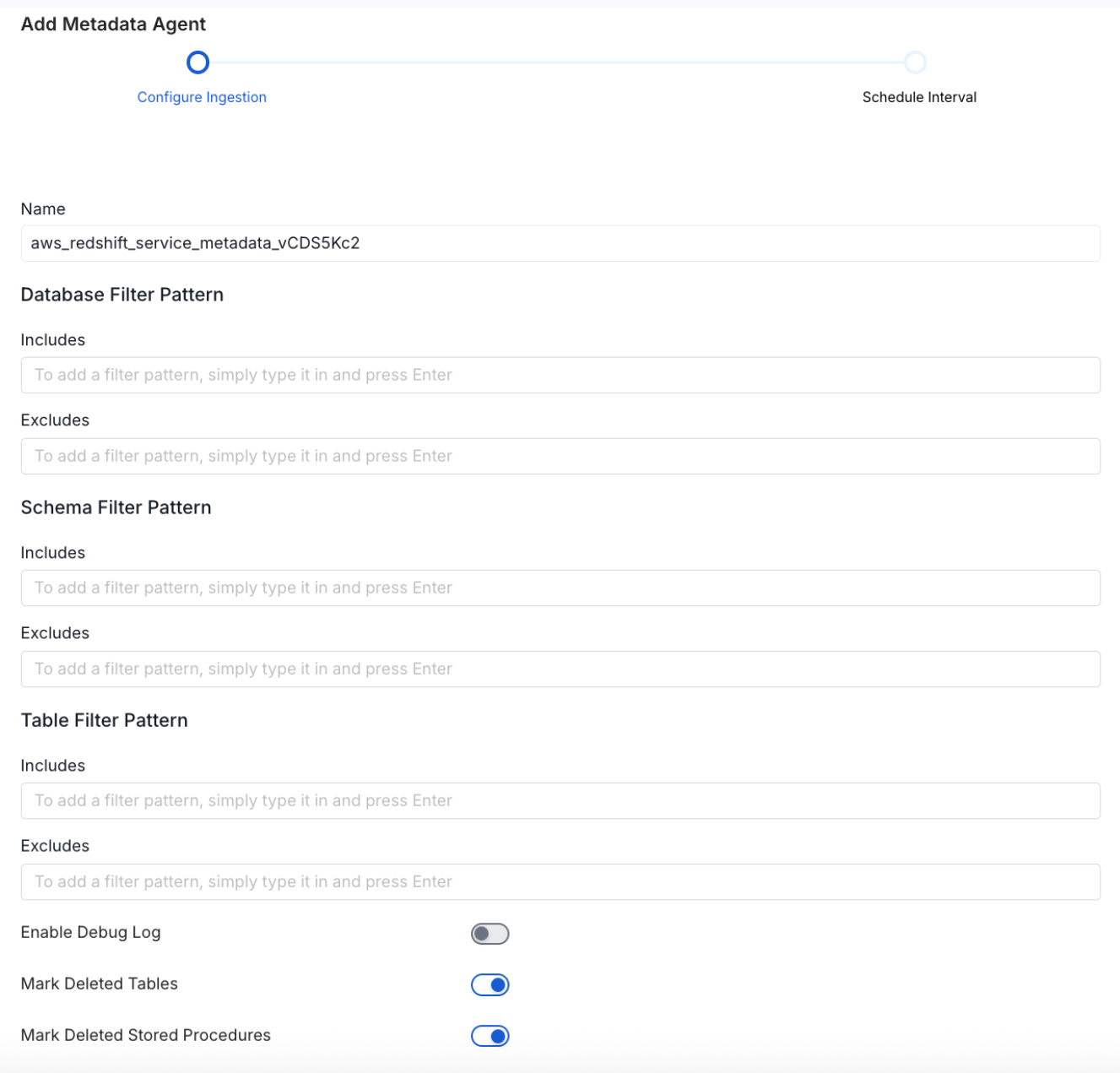
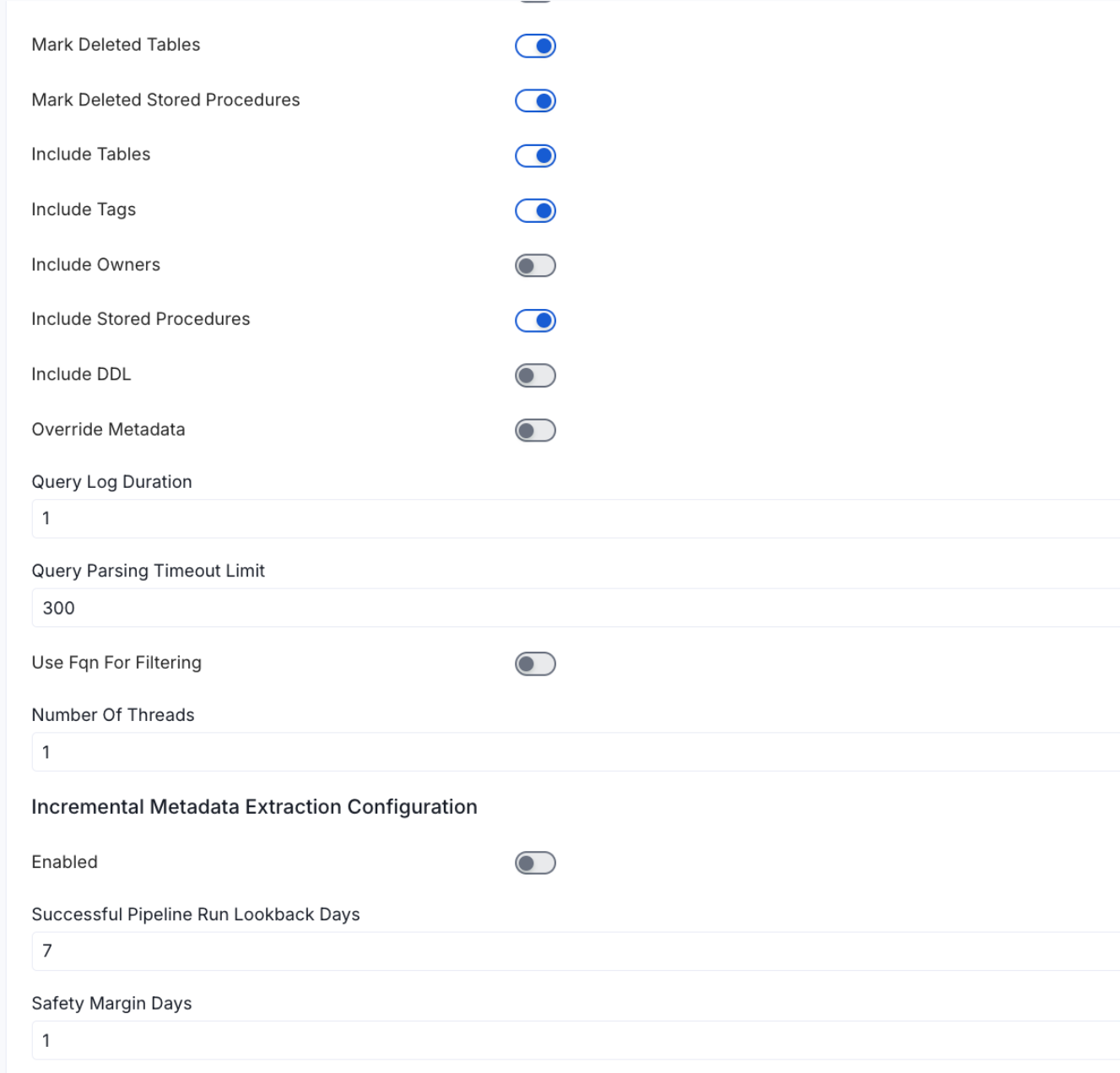


Metadata Ingestion Options
- Name: This field refers to the name of ingestion pipeline, you can customize the name or use the generated name.
-
Database Filter Pattern (Optional): Use to database filter patterns to control whether or not to include database as part of metadata ingestion.
- Include: Explicitly include databases by adding a list of comma-separated regular expressions to the Include field. OpenMetadata will include all databases with names matching one or more of the supplied regular expressions. All other databases will be excluded.
- Exclude: Explicitly exclude databases by adding a list of comma-separated regular expressions to the Exclude field. OpenMetadata will exclude all databases with names matching one or more of the supplied regular expressions. All other databases will be included.
-
Schema Filter Pattern (Optional): Use to schema filter patterns to control whether to include schemas as part of metadata ingestion.
- Include: Explicitly include schemas by adding a list of comma-separated regular expressions to the Include field. OpenMetadata will include all schemas with names matching one or more of the supplied regular expressions. All other schemas will be excluded.
- Exclude: Explicitly exclude schemas by adding a list of comma-separated regular expressions to the Exclude field. OpenMetadata will exclude all schemas with names matching one or more of the supplied regular expressions. All other schemas will be included.
-
Table Filter Pattern (Optional): Use to table filter patterns to control whether to include tables as part of metadata ingestion.
- Include: Explicitly include tables by adding a list of comma-separated regular expressions to the Include field. OpenMetadata will include all tables with names matching one or more of the supplied regular expressions. All other tables will be excluded.
- Exclude: Explicitly exclude tables by adding a list of comma-separated regular expressions to the Exclude field. OpenMetadata will exclude all tables with names matching one or more of the supplied regular expressions. All other tables will be included.
- Enable Debug Log (toggle): Set the Enable Debug Log toggle to set the default log level to debug.
- Mark Deleted Tables (toggle): Set the Mark Deleted Tables toggle to flag tables as soft-deleted if they are not present anymore in the source system.
- Mark Deleted Tables from Filter Only (toggle): Set the Mark Deleted Tables from Filter Only toggle to flag tables as soft-deleted if they are not present anymore within the filtered schema or database only. This flag is useful when you have more than one ingestion pipelines. For example if you have a schema
- includeTables (toggle): Optional configuration to turn off fetching metadata for tables.
- includeViews (toggle): Set the Include views toggle to control whether to include views as part of metadata ingestion.
- includeTags (toggle): Set the ‘Include Tags’ toggle to control whether to include tags as part of metadata ingestion.
- includeOwners (toggle): Set the ‘Include Owners’ toggle to control whether to include owners to the ingested entity if the owner email matches with a user stored in the OM server as part of metadata ingestion. If the ingested entity already exists and has an owner, the owner will not be overwritten.
- includeStoredProcedures (toggle): Optional configuration to toggle the Stored Procedures ingestion.
- includeDDL (toggle): Optional configuration to toggle the DDL Statements ingestion.
- queryLogDuration (Optional): Configuration to tune how far we want to look back in query logs to process Stored Procedures results.
- queryParsingTimeoutLimit (Optional): Configuration to set the timeout for parsing the query in seconds.
- useFqnForFiltering (toggle): Regex will be applied on fully qualified name (e.g service_name.db_name.schema_name.table_name) instead of raw name (e.g. table_name).
-
Incremental (Beta): Use Incremental Metadata Extraction after the first execution. This is done by getting the changed tables instead of all of them. Only Available for BigQuery, Redshift and Snowflake
- Enabled: If
True, enables Metadata Extraction to be Incremental. - lookback Days: Number of days to search back for a successful pipeline run. The timestamp of the last found successful pipeline run will be used as a base to search for updated entities.
- Safety Margin Days: Number of days to add to the last successful pipeline run timestamp to search for updated entities.
- Enabled: If
- Threads (Beta): Use a Multithread approach for Metadata Extraction. You can define here the number of threads you would like to run concurrently.
Schedule the Ingestion and Deploy
Scheduling can be set up at an hourly, daily, weekly, or manual cadence. The
timezone is in UTC. Select a Start Date to schedule for ingestion. It is
optional to add an End Date.Review your configuration settings. If they match what you intended,
click Deploy to create the service and schedule metadata ingestion.If something doesn’t look right, click the Back button to return to the
appropriate step and change the settings as needed.After configuring the workflow, you can click on Deploy to create the
pipeline.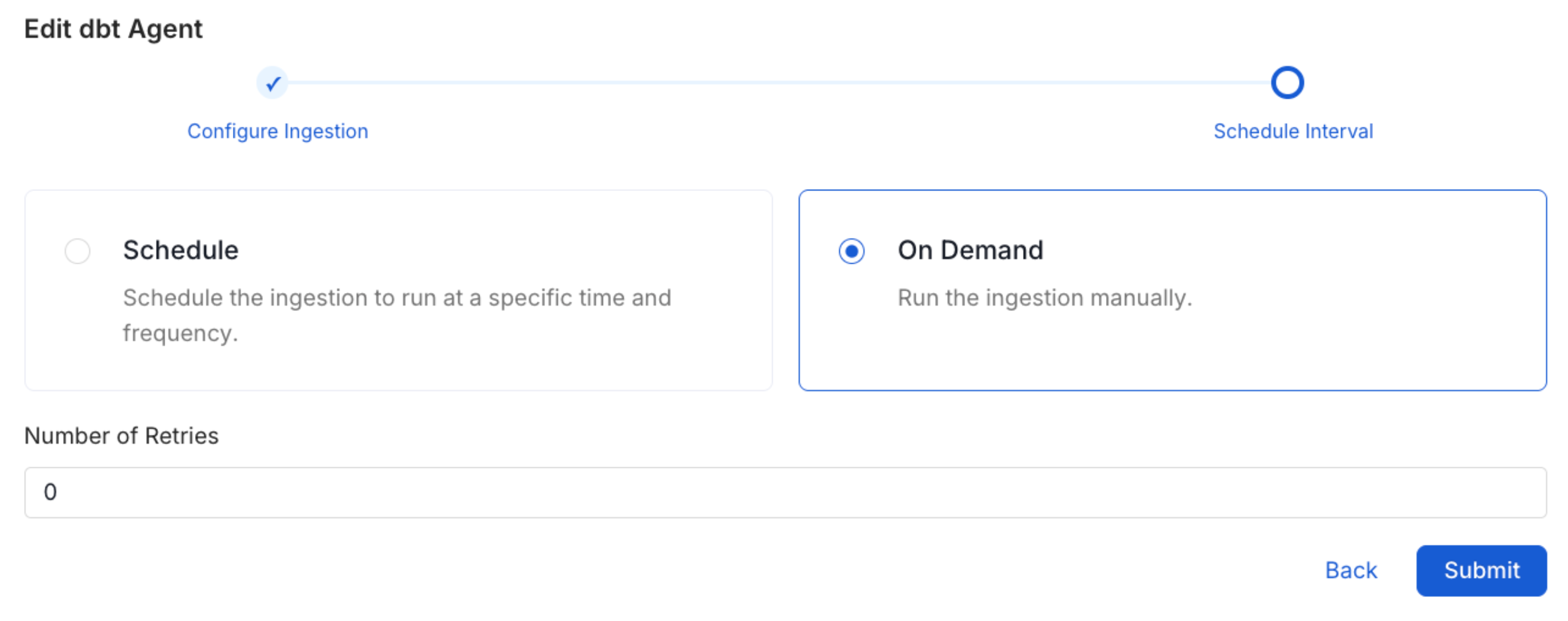

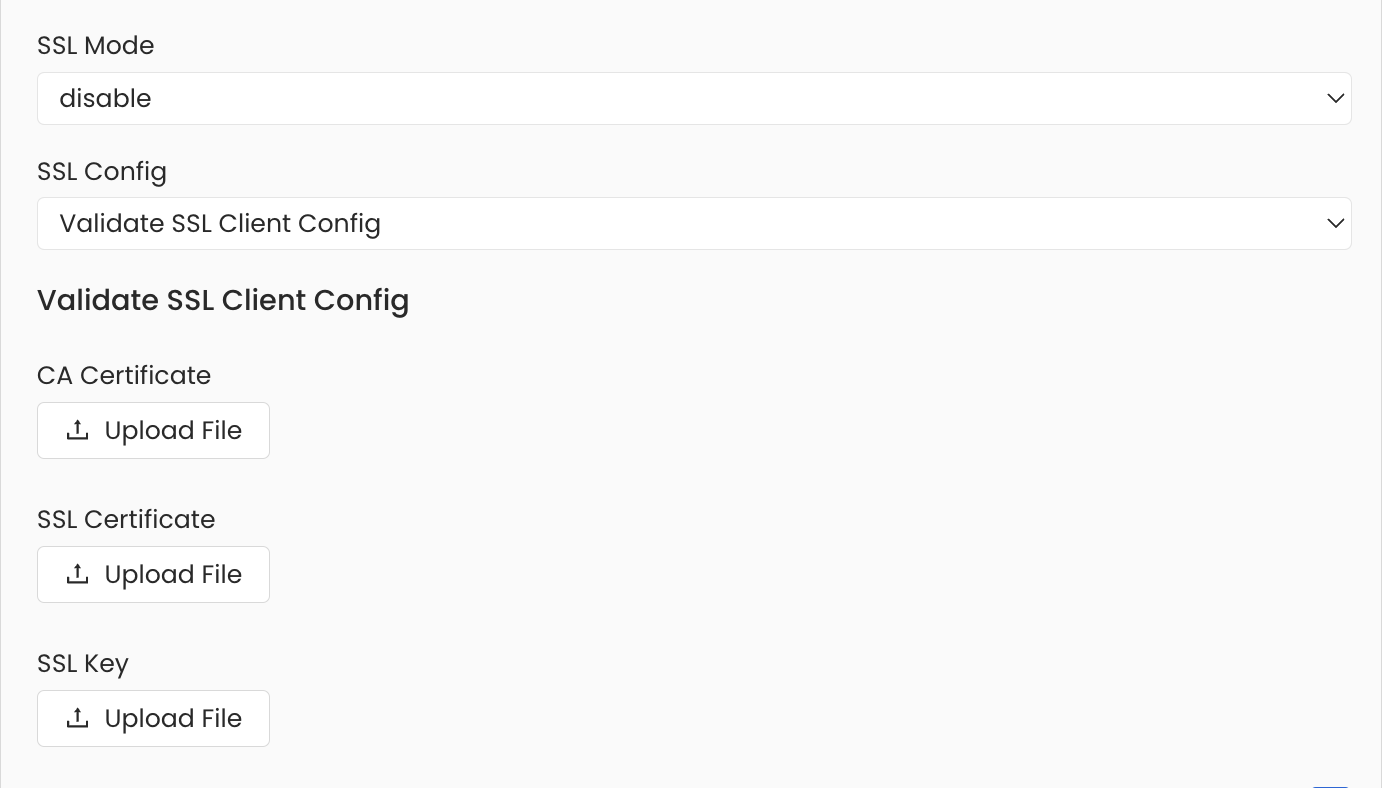
Securing Redshift Connection with SSL in OpenMetadata
To establish secure connections between OpenMetadata and a Redshift database, you can configure SSL using different SSL modes provided by Redshift, each offering varying levels of security. UnderAdvanced Config, specify the SSL mode appropriate for your connection, such as prefer, verify-ca, allow, and others. After selecting the SSL mode, provide the CA certificate used for SSL validation (caCertificate). Note that Redshift requires only the CA certificate for SSL validation.
Reverse Metadata
Description Management
Redshift supports description updates at all levels:- Database level
- Schema level
- Table level
- Column level
Owner Management
Redshift supports owner management at the following levels:- Database level
- Schema level
- Table level
Tag Management
❌ Tag management is not supported for Redshift.Custom SQL Template
Redshift supports custom SQL templates for metadata changes. The template is interpreted using python f-strings.Here are examples of custom SQL queries for metadata changes:Requirements for Reverse Metadata
In addition to the basic ingestion requirements, for reverse metadata ingestion the user needs:SUPERUSERprivilege to the user
Related
Usage Workflow
Learn more about how to configure the Usage Workflow to ingest Query information from the UI.
Lineage Workflow
Learn more about how to configure the Lineage from the UI.
Profiler Workflow
Learn more about how to configure the Data Profiler from the UI.
Data Quality Workflow
Learn more about how to configure the Data Quality tests from the UI.
dbt Integration
Learn more about how to ingest dbt models’ definitions and their lineage.